Rosenzweig-MacArthur Predator-Prey model
Source:vignettes/rm_predator_prey.Rmd
rm_predator_prey.RmdRosenzweig-MacArthur predator-prey model (Pineda-Krch et al. 2007).
dN/dt = r(1-N/K - alpha/(1+wN))NP
dP/dt = c*alpha/(1+wN))NPThis model has five reactions with the following per capita rates,
prey birth: b
prey death: d+(b-d)N/K
predation: alpha/(1+wN)
predator birth: c*alpha/(1+wN)N
predator death: gPropensity functions:
a1 = b * N
a2 = (d+(b-d)N/K) * N
a3 = alpha/(1+wN) * N * P
a4 = c*alpha/(1+wN) * N * P
a5 = g * PDefine parameters
library(GillespieSSA2)
sim_name <- "Rosenzweig-MacArthur Predator-Prey model"
params <- c(
b = 2,
d = 1,
K = 1000,
alpha = 0.005,
w = 0.0025,
c = 2,
g = 2
)
final_time <- 10
initial_state <- c(N = 500, P = 500)Define reactions
reactions <- list(
reaction("b * N", c(N = +1)),
reaction("(d + (b - d) * N / K) * N", c(N = -1)),
reaction("alpha / (1 + w * N) * N * P", c(N = -1)),
reaction("c * alpha / ( 1 + w * N) * N * P", c(P = +1)),
reaction("g * P", c(P = -1))
)Run simulations with the Exact method
set.seed(1)
out <- ssa(
initial_state = initial_state,
reactions = reactions,
params = params,
final_time = final_time,
method = ssa_exact(),
sim_name = sim_name
)
plot_ssa(out)## Loading required namespace: ggplot2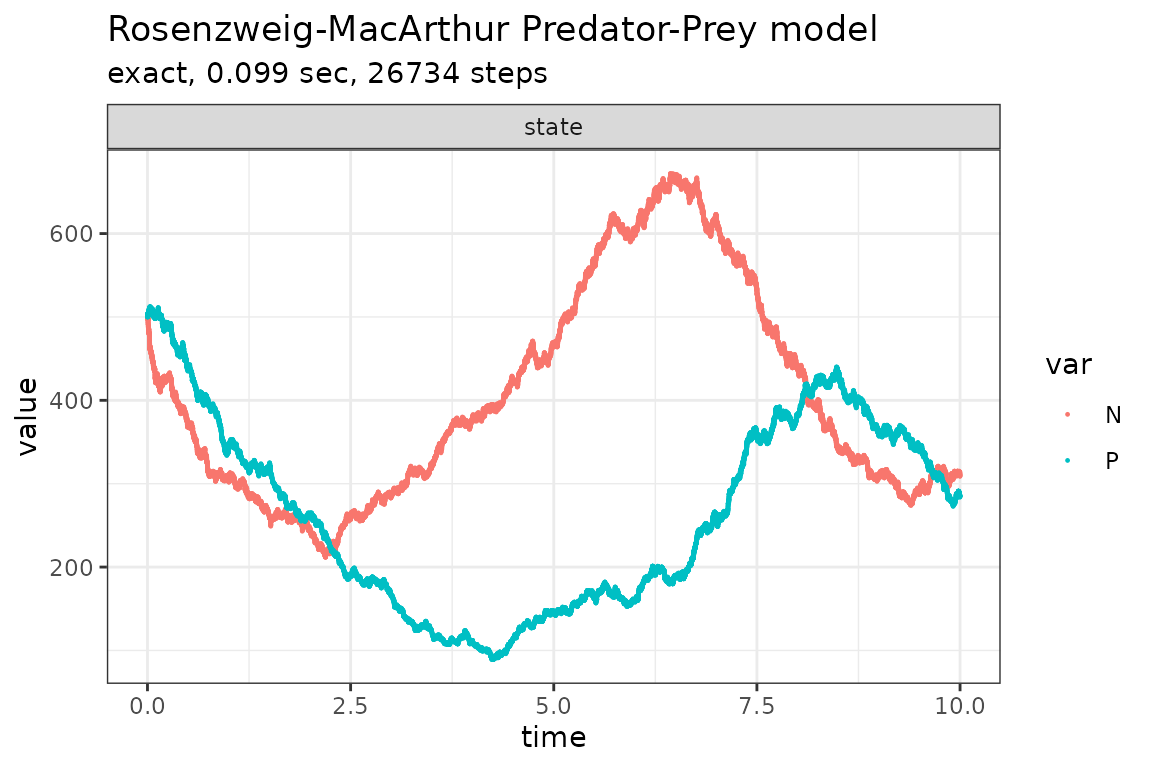
Run simulations with the Explict tau-leap method
set.seed(1)
out <- ssa(
initial_state = initial_state,
reactions = reactions,
params = params,
final_time = final_time,
method = ssa_etl(tau = .01),
sim_name = sim_name
)
plot_ssa(out)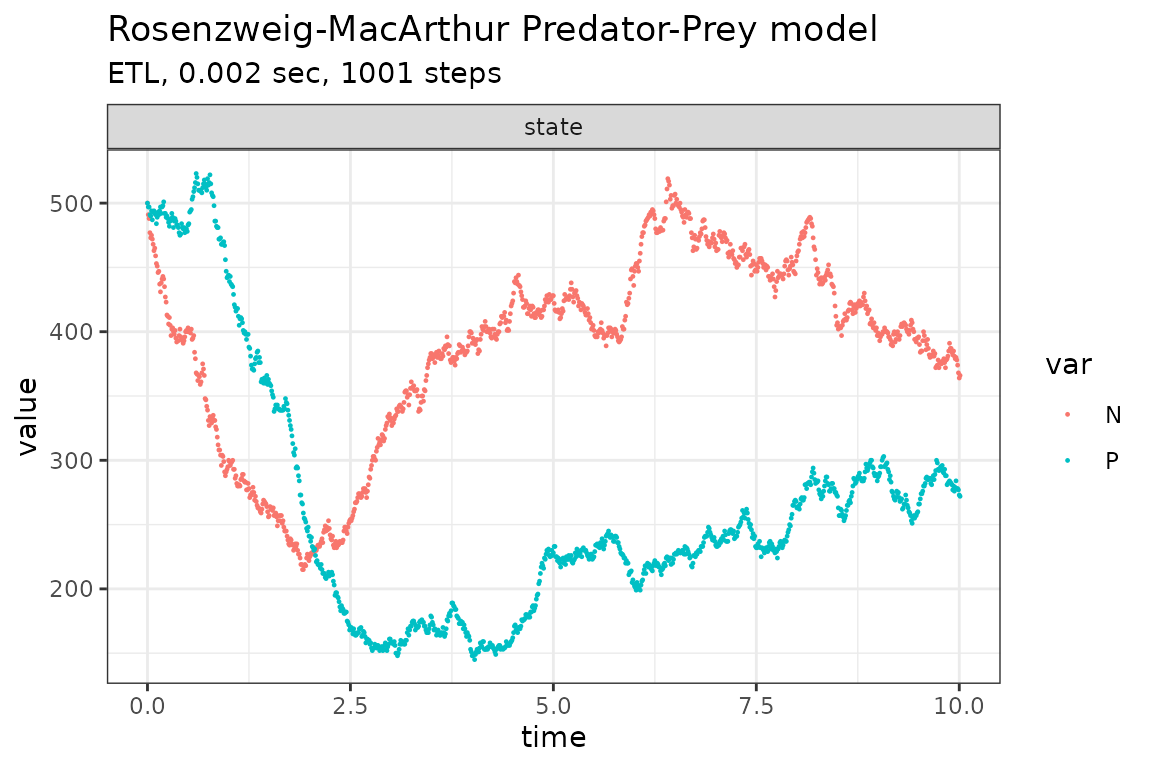
Run simulations with the Binomial tau-leap method
set.seed(1)
out <- ssa(
initial_state = initial_state,
reactions = reactions,
params = params,
final_time = final_time,
method = ssa_btl(),
sim_name = sim_name
)
plot_ssa(out)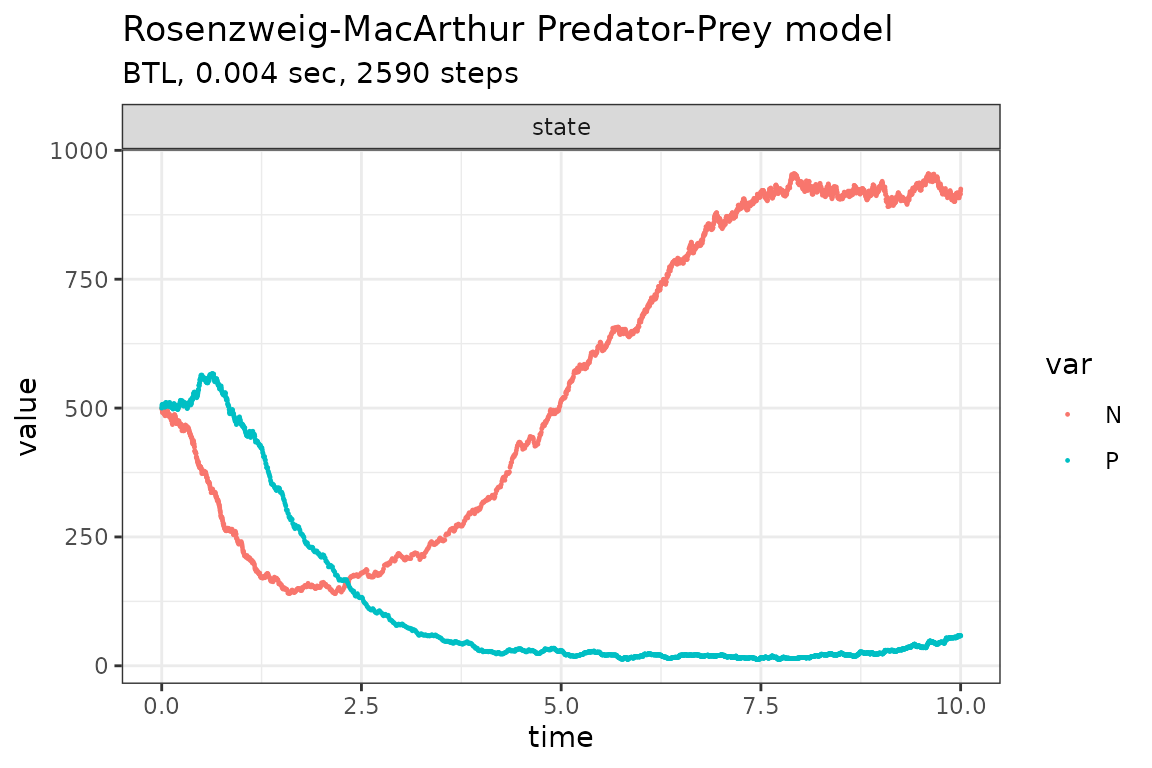
References
Pineda-Krch, Mario, Hendrik J. Blok, Ulf Dieckmann, and Michael Doebeli.
2007. “A Tale of Two Cycles – Distinguishing Quasi-Cycles and
Limit Cycles in Finite Predator–Prey Populations.” Oikos
116 (1): 53–64. https://doi.org/https://doi.org/10.1111/j.2006.0030-1299.14940.x.